Cookbook SMRT Link Webservices using pbcommand¶
In [2]:
# https://github.com/PacificBiosciences/pbcommand
# install via
# pip install -e git://github.com/PacificBiosciences/pbcommand.git#egg=pbcommand`
import pbcommand
from pbcommand.services import ServiceAccessLayer
print pbcommand.get_version()
0.6.0
Configure and Get Status¶
In [3]:
host = "smrtlink-beta"
port = 8081
sal = ServiceAccessLayer(host, port)
In [4]:
sal.get_status()
Out[4]:
{u'id': u'smrtlink_analysis',
u'message': u'Services have been up for 257 hours, 10 minutes and 1.427 second.',
u'uptime': 925801427,
u'user': u'secondarytest',
u'uuid': u'6742aebc-bce8-45b6-acfe-422c9d941884',
u'version': u'0.3.0-bbcc1b5'}
Get SubreadSet from SMRT Link Services¶
Get SubreadSet details by UUID¶
In [5]:
# Example SubreadSet
subreadset_uuid = "4463c7d8-4b6e-43a3-93ff-325628066a0b"
In [6]:
ds = sal.get_subreadset_by_id(subreadset_uuid)
In [7]:
ds['metadataContextId'], ds['name'], ds['id'], ds['jobId'], ds['wellName'], ds['path'], ds['uuid']
Out[7]:
(u'm54008_160614_021525',
u'SMS_FleaBenchmark_15kbEcoli_trypsin_A8_061316',
19251,
12469,
u'A01',
u'/pbi/collections/315/3150259/r54008_20160614_021104/1_A01/m54008_160614_021525.subreadset.xml',
u'4463c7d8-4b6e-43a3-93ff-325628066a0b')
In [9]:
# or get by Int id
dx = sal.get_subreadset_by_id(ds['id'])
dx['uuid'], ds['uuid']
Out[9]:
(u'4463c7d8-4b6e-43a3-93ff-325628066a0b',
u'4463c7d8-4b6e-43a3-93ff-325628066a0b')
In [10]:
# Get the Import Job associated with the SubreadSet. This Job computes the metrics at import time
In [11]:
import_job_id = ds['jobId']
Get SubreadSet Import Job and Import Report Metrics¶
In [12]:
import_job = sal.get_import_job_by_id(import_job_id)
In [13]:
import_job
Out[13]:
<ServiceJob i:12469 state: SUCCESSFUL created:06-14-2016 05:45.46 by:kgleitsman name:Job import-data... runtime: 0.00 sec >
In [14]:
import_job.state
Out[14]:
'SUCCESSFUL'
In [15]:
import_job.name
Out[15]:
'Job import-dataset'
In [16]:
import_report_attributes = sal.get_import_job_report_attrs(import_job.id)
In [17]:
import_report_attributes # This are what are displayed in SMRT Link for Import Job Types
Out[17]:
{u'adapter_xml_report.adapter_dimers': 0.02,
u'adapter_xml_report.short_inserts': 1.63,
u'raw_data_report.insert_length': 5974,
u'raw_data_report.nbases': 2769788750,
u'raw_data_report.nreads': 539045,
u'raw_data_report.read_length': 5138,
u'raw_data_report.read_n50': 10250}
Get Analysis (pbsmrtpipe) Job¶
In [18]:
analysis_job_id = 12524 # Random Example Job
In [19]:
analysis_job = sal.get_analysis_job_by_id(analysis_job_id)
In [20]:
analysis_job
Out[20]:
<ServiceJob i:12524 state: SUCCESSFUL created:06-15-2016 01:02.30 by:avnguyen name:54082_FAT1_2Cel... runtime: 0.00 sec >
In [21]:
analysis_job.settings['pipelineId']
Out[21]:
u'pbsmrtpipe.pipelines.sa3_sat'
Get the Entry Points to the Pipeline¶
In [22]:
# Get Entry Points Use
eps = sal.get_analysis_job_entry_points(analysis_job_id)
In [23]:
for e in eps:
print e
JobEntryPoint(job_id=12524, dataset_uuid=u'3adaa998-8ba9-4f7c-97c4-2ef343b419bb', dataset_metatype=u'PacBio.DataSet.SubreadSet')
JobEntryPoint(job_id=12524, dataset_uuid=u'c2f73d21-b3bf-4b69-a3a3-6beeb7b22e26', dataset_metatype=u'PacBio.DataSet.ReferenceSet')
Get Report Metrics for Analysis Job¶
In [24]:
analysis_report_attributes = sal.get_analysis_job_report_attrs(analysis_job_id)
In [25]:
analysis_report_attributes
Out[25]:
{u'coverage.depth_coverage_mean': 147.6752298049565,
u'coverage.missing_bases_pct': 0.0,
u'mapping_stats.mapped_readlength_max': 16470,
u'mapping_stats.mapped_readlength_mean': 3743,
u'mapping_stats.mapped_readlength_n50': 5974,
u'mapping_stats.mapped_readlength_q95': 10400,
u'mapping_stats.mapped_reads_n': 2319,
u'mapping_stats.mapped_subread_bases_n': 7654453,
u'mapping_stats.mapped_subread_concordance_mean': 0.8006,
u'mapping_stats.mapped_subread_readlength_max': 4339.0,
u'mapping_stats.mapped_subread_readlength_mean': 1055,
u'mapping_stats.mapped_subreadlength_n50': 1328,
u'mapping_stats.mapped_subreadlength_q95': 2070,
u'mapping_stats.mapped_subreads_n': 7257,
u'sat.accuracy': 1.0,
u'sat.coverage': 1.0,
u'sat.instrument': u'54082',
u'sat.mapped_readlength_mean': 3743,
u'sat.reads_in_cell': 2319,
u'variants.longest_contig_name': u'lambda_NEB3011',
u'variants.mean_contig_length': 48502.0,
u'variants.weighted_mean_bases_called': 1.0,
u'variants.weighted_mean_concordance': 1.0,
u'variants.weighted_mean_coverage': 147.6752298049565}
Example of Getting Mulitple Jobs and extracting Metrics to build a Table¶
In [26]:
# Grab some example jobs
job_ids = [12357, 12359, 12360, 12361, 12362, 12367]
In [27]:
def get_attrs_from_job_id(sal_, job_id_):
d = sal_.get_analysis_job_report_attrs(job_id_)
d['job_id'] = job_id_
d['host'] = host
d['port'] = port
return d
In [28]:
datum = [get_attrs_from_job_id(sal, x) for x in job_ids]
In [29]:
# Plot some data
import matplotlib.pyplot as plt
import matplotlib
import pandas as pd
import seaborn as sns
sns.set_style("whitegrid")
%matplotlib inline
In [30]:
df = pd.DataFrame(datum)
In [31]:
df
Out[31]:
| coverage.depth_coverage_mean | coverage.missing_bases_pct | host | job_id | mapping_stats.mapped_readlength_max | mapping_stats.mapped_readlength_mean | mapping_stats.mapped_readlength_n50 | mapping_stats.mapped_readlength_q95 | mapping_stats.mapped_reads_n | mapping_stats.mapped_subread_bases_n | ... | mapping_stats.mapped_subread_readlength_mean | mapping_stats.mapped_subreadlength_n50 | mapping_stats.mapped_subreadlength_q95 | mapping_stats.mapped_subreads_n | port | variants.longest_contig_name | variants.mean_contig_length | variants.weighted_mean_bases_called | variants.weighted_mean_concordance | variants.weighted_mean_coverage | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
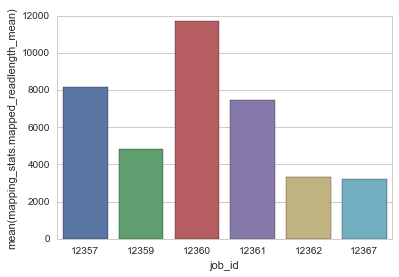
| 0 | 40288.305513 | 0.0 | smrtlink-beta | 12357 | 35886 | 8178 | 16148 | 22480 | 272393 | 2074235751 | ... | 1772 | 2075 | 2880 | 1170748 | 8081 | lambda_NEB3011 | 48502.0 | 1.0 | 1.000000 | 40288.305513 |
| 1 | 364.705681 | 0.0 | smrtlink-beta | 12359 | 41461 | 4810 | 8968 | 16530 | 396370 | 1888916069 | ... | 3732 | 6039 | 10010 | 506093 | 8081 | ecoliK12_pbi_March2013 | 4642522.0 | 1.0 | 0.999834 | 364.705681 |
| 2 | 91904.489318 | 0.0 | smrtlink-beta | 12360 | 40505 | 11687 | 20660 | 30290 | 17762 | 191479775 | ... | 1673 | 2016 | 2200 | 114447 | 8081 | tkb_v1_C5_insertOnly | 1966.0 | 1.0 | 1.000000 | 91904.489318 |
| 3 | 59869.400293 | 0.0 | smrtlink-beta | 12361 | 32517 | 7470 | 13719 | 20370 | 449931 | 3109535638 | ... | 1872 | 2262 | 3150 | 1660986 | 8081 | lambda_NEB3011 | 48502.0 | 1.0 | 1.000000 | 59869.400293 |
| 4 | 156.527813 | 0.0 | smrtlink-beta | 12362 | 42024 | 3304 | 5924 | 11470 | 248365 | 814056472 | ... | 2859 | 4536 | 8850 | 284695 | 8081 | ecoliK12_pbi_March2013 | 4642522.0 | 1.0 | 0.999937 | 156.527813 |
| 5 | 257.757219 | 0.0 | smrtlink-beta | 12367 | 37148 | 3237 | 5708 | 10240 | 413560 | 1330448079 | ... | 2827 | 4643 | 8460 | 470554 | 8081 | ecoliK12_pbi_March2013 | 4642522.0 | 1.0 | 0.999829 | 257.757219 |
6 rows × 22 columns
In [32]:
sns.barplot(x="job_id", y="mapping_stats.mapped_readlength_mean", data=df)
Out[32]:
<matplotlib.axes._subplots.AxesSubplot at 0x10bd75b90>
In [ ]: